-
Behaviour Scoring
-
- 1. Anxious in unfamiliar situations
- 2. Fear Of Noises
- 3. Fear of Novel Objects
- 4. Fear of Underfootings
- 5. Fear of Dogs
- 6. Fear of Stairs
- 7. Fear of Traffic
- 8. Separation Anxiety
- 9. Hyper-Attachment
- 10. Fear Of Strangers
- 11. Body Handling Concern
- 12. Retreats When Reached For
- 13. Harness Handle On Back Sensitivity
- 14. Avoidance Of Blowing Fan
- 15. Body Sensitivity To Object Contact
- 16. Anxious About Riding In Vehicles
- 17. Inhibited or passively avoidant when exposed to potentially stressful situations
- 18. Activated when exposed to potentially stressful situations
- 19. Excitable
- 20. Slow To Return To Productive Emotional State
- 21. Fidgety When Handler Is Idle
- 22. Fear On Elevated Areas, Drop-Offs Etc.
- 23. Barks Persistently
- 24. High Energy Level
- 25. Lacks Focus
- 26. Movement Excites
- 27. Chasing Animals
- 28. Dog Distraction
- 29. Sniffing
- 30. Scavenges
- 31. Inappropriate Behavior Around The Home
- 32. Lacks Initiative
- 33. Not Willing
- 34. Resource Guarding Toward People
- 35. Aggression Toward Strangers
- 36. Aggression Toward Dogs
- 37. Resource Guarding Toward Dogs Or Other Pets
- 38. Inappropriate Elimination While Working En Route
- 39. Socially Inappropriate Behavior With People
- 40. Inconsistent
- 41. Handler/Dog Team
- 42. Relationship Skills
- 43. Comparison 9 To 1 Score
- 44. Socially Inappropriate Behavior With Dogs
- 45. Thunder Reaction Prior To, During Or Immediately After A Thunderstorm
- 46. Kennels Poorly
- 47. Working Speed
- 48. Gait When Moving Out
- 49. Housebreaking Problems
- 50. Innate Desire To Work
- 51. Avoidance Of Exhaust From Vehicles
- Show all articles ( 36 ) Collapse Articles
-
-
-
Practice Videos
-
Behavior Testing
-
Database User Manual
-
-
-
- Adding a New Dog (using Manage Your Dog’s Data, MyDogs)
- Alerts
- BCL, Behavior Checklist
- Elbow Quick, Add new
- Estrus & Whelps
- Eye Quick
- Genetic Test Panel
- Genetic Test Quick
- Health Diagnoses Add/Edit
- Health History Report
- Health Normals, Add new
- Heart Quick
- Hip BVA, Add new
- Hip FCI, Add new
- Hip OFA, Add new
- Hip Penn Hip, Add new
- Photos PDFs etc.
- Private Notes
- Procedures, Add new
- Reminders
- Share my dog data to another organization
- Skin Quick
- Status History
- Weight - Entering a dog's weight
- ADI Public Access Test
- Hip Vezzoni, Add new
- Status Detail
- Edit or Change Call Name / Pedigree Name / Owner ID
- Add New Microchip / Delete Incorrect Microchip
- End Reasons
- Juvenile Estrus
- Communications Activities
- Incidents
- Show all articles ( 18 ) Collapse Articles
-
- Articles coming soon
-
-
-
-
- Alternate Therapy/Rehab
- Diagnostic Imaging, Add new / Edit or Delete
- Diet
- Elbow Quick, Add new / Edit or Delete
- Estrus & Whelps, Add new
- Eye Quick, Add new / Edit or Delete
- Genetic Test Quick, Add new / Edit or Delete
- Health Diagnoses, Add new / Edit (Update) or Delete
- Health History Report, Generate a PDF
- Health Normals
- Health Screening List
- Hip OFA Add new / Edit or Delete
- Hip Penn Hip Add new / Edit or Delete
- Hospitalization, Add new
- Kennel Tasks, Add new / Edit or Delete
- Lab, Add new / Edit or Delete
- Photos, PDFs, etc., Add new
- Reminders Add new / Edit or Delete
- Rx, Add new / Edit or Delete
- Semen Cryo, Add new / Edit or Delete
- Skin Quick Add new / Edit or Delete
- SOAP, Add new / Edit or Delete
- Status History
- Supplies Used, Add new / Edit or Delete
- Surgery, Add new
- Treatments Add new / Edit or Delete
- Vaccines Add new / Edit or Delete
- Weight and BCS Body Condition Score - Add new / Edit or Delete
- Hip Vezzoni, Add new
- Show all articles ( 14 ) Collapse Articles
-
-
-
-
Early Socialization
-
- Video - Coat Desensitization
- Video - Novel Objects
- Video - Trolley Ride with Mom
- Early Puppy Socialization - Novel Objects video
- Early Puppy Socialization – Novel Sounds video
- Early Puppy Socialization – Introducing New Environments video
- Early Puppy Socialization – Motor Development, Balance, Coordination, Proprioception video
- Early Puppy Socialization – Passive Environmental Enrichment in the Den video
- Early Puppy Socialization – Stairs
-
-
Genetic Selection & Inbreeding
-
- What are EBVs and how do they help?
- How EBVs are calculated
- What is needed to calculate EBVs and EBV accuracy?
- Using EBVs effectively
- Selection index
- Why are EBVs different for littermates?
- Presentation Recording: Improving behavior using EBVs
- Presentation Recording: Using EBVs successfully
- Presentation Recording - Improving health using EBVs
-
Webinars
-
Reproduction
-
Organization Management
How EBVs are calculated
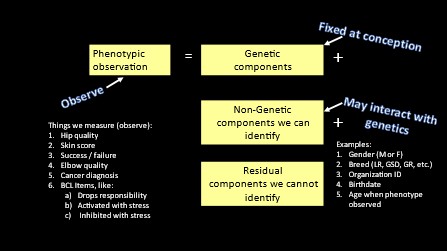
EBVs are calculated from special quantitative genetics software using data on the phenotypes of all relatives and their pedigree relationships. The data used in the calculation includes the trait of interest such as elbow dysplasia measure from all or most dogs and other measures that influence variation in the expression of the trait such as sex, scorer, age of dog when measured, etc.
These calculations do not require knowing the actual genes involved. However it is also possible to incorporate into the statistical calculations genomic data that identify specific genes or markers that underlie variation in quantitative traits. The complex mathematical calculations can be done for traits with a continuous range of values like height and weight and also for threshold traits which have discrete values such as severity of elbow dysplasia or even binary traits.
Quantitative traits are often polygenic meaning they are influenced by many and often a large number of genes, each having an effect. They are also affected by environmental influences.
In livestock breeding, genomic data are also included in the genetic model. EBVs that are calculated using genomic data along with phenotype data are called genomically enhanced breeding values or gEBVs. Genomically enhanced breeding values are not currently in use with canine breeding programs.
The steps are to:
1. Combine the data and pedigree relationships for all dogs that have measures for the trait of a particular breed or in some cases multiple breeds. IWDR ties all dogs together and provides structured data entry. Lots of data are needed.
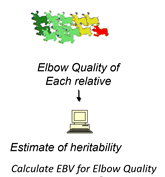
2. Estimate the heritability to determine if the trait is sufficiently heritable to respond to genetic selection.
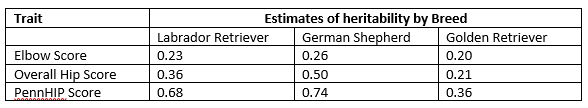
Heritability (h2) is officially defined as the proportion of total variance observed in a trait that arises from genetic differences among animals. Variance measures the degree to which individual measurements depart from the population mean. In simpler terms, heritability is the proportion of the parental superiority over the population average that parents transmit to their offspring. Even more simply, it is the degree to which genes cause the phenotype that can be measured. Because it is a ratio of a part of something to all of something, it must be a number between 0 and 1. It can be estimated in several different ways, but to do the estimation correctly requires training. Most agricultural universities employ trained scientists known as animal breeders who possess this training. The higher the heritability, the greater and faster genetic selection can improve the trait.
Examples of heritability for IWDR data:
The genetic model which can be thought of as a list of variables that includes the phenotype and other information called ‘non-genetic components’ that can be identified. Non-genetic components are bits of data that have been tested statistically to influence the differences in the scores for the trait being studied. Examples may include sex, age that the phenotype was measured, organization where measurement was scored. The data are run on specialized software using the configuration of the genetic model and the estimate of heritability provided by the software.
3. Calculate the EBVs – Estimated Breeding Values can be calculated for traits with high enough estimates of heritability. EBVs need to recalculated regularly because new data, especially on the dog and close relatives, may change a dog’s ranking for better or worse. IWDR updates the EBVs every week.
